Publications
Most up to date paper list on Google Scholar
Analysis of developmental gene expression using smFISH and in silico staging of C. elegans embryos
Authors: Laura Breimann, Ella Bahry, Marwan Zouinkhi, Klim Kolyvanov, Lena Annika Street, Stephan Preibisch and Sevinç Ercan
Date: May 16, 2024
Journal: bioRxiv
Abstract: Regulation of transcription during embryogenesis is key to development and differentiation. To study transcript expression throughout Caenorhabditis elegans embryogenesis at single-molecule resolution, we developed a high-throughput single-molecule fluorescence in situ hybridization (smFISH) method that relies on computational methods to developmentally stage embryos and quantify individual mRNA molecules in single embryos. We applied our system to sdc-2, a zygotically transcribed gene essential for hermaphrodite development and dosage compensation. We found that sdc-2 is rapidly activated during early embryogenesis by increasing both the number of mRNAs produced per transcription site and the frequency of sites engaged in transcription. Knockdown of sdc-2 and dpy-27, a subunit of the dosage compensation complex (DCC), increased the number of active transcription sites for the X chromosomal gene dpy-23 but not the autosomal gene mdh-1, suggesting that the DCC reduces the frequency of dpy-23 transcription. The temporal resolution from in silico staging of embryos showed that the deletion of a single DCC recruitment element near the dpy-23 gene causes higher dpy-23 mRNA expression after the start of dosage compensation, which could not be resolved using mRNAseq from mixed-stage embryos. In summary, we have established a computational approach to quantify temporal regulation of transcription throughout C. elegans embryogenesis and demonstrated its potential to provide new insights into developmental gene regulation.
Link: Read Paper
Cryosectioning-enabled super-resolution microscopy for studying nuclear architecture at the single protein level
Authors: Johannes Stein, Maria Ericsson, Michel Nofal, Lorenzo Magni, Sarah Aufmkolk, Ryan B. McMillan, Laura Breimann, Conor P. Herlihy, S. Dean Lee, Andréa Willemin, Jens Wohlmann, Laura Arguedas-Jimenez, Peng Yin, Ana Pombo, George M. Church, Chao-ting Wu
Date: Feburary 5, 2024
Journal: bioRxiv
Abstract: DNA-PAINT combined with total Internal Reflection Fluorescence (TIRF) microscopy enables the highest localization precisions, down to single nanometers in thin biological samples, due to TIRF’s unique method for optical sectioning and attaining high contrast. However, most cellular targets elude the accessible TIRF range close to the cover glass and thus require alternative imaging conditions, affecting resolution and image quality. Here, we address this limitation by applying ultrathin physical cryosectioning in combination with DNA-PAINT. With “tomographic & kinetically-enhanced” DNA-PAINT (tokPAINT), we demonstrate the imaging of nuclear proteins with sub-3 nanometer localization precision, advancing the quantitative study of nuclear organization within fixed cells and mouse tissues at the level of single antibodies. We believe that ultrathin sectioning combined with the versatility and multiplexing capabilities of DNA-PAINT will be a powerful addition to the toolbox of quantitative DNA-based super-resolution microscopy in intracellular structural analyses of proteins, RNA and DNA in situ.
Link: Read Paper
mRNA stability and m6A are major determinants of subcellular mRNA localization in neurons
Authors: Inga Loedige, Artem Baranovskii, Samantha Mendonsa, Sayaka Dantsuji, Niko Popitsch, Laura Breimann, Nadja Zerna, Vsevolod Cherepanov, Miha Milek, Stefan Ameres, Marina Chekulaeva
Date: August 3, 2023
Journal: Molecular Cell
Abstract: For cells to perform their biological functions, they need to adopt specific shapes and form functionally distinct subcellular compartments. This is achieved in part via an asymmetric distribution of mRNAs within cells. Currently, the main model of mRNA localization involves specific sequences called “zipcodes” that direct mRNAs to their proper locations. However, while thousands of mRNAs localize within cells, only a few zipcodes have been identified, suggesting that additional mechanisms contribute to localization. Here, we assess the role of mRNA stability in localization by combining the isolation of the soma and neurites of mouse primary cortical and mESC-derived neurons, SLAM-seq, m6A-RIP-seq, the perturbation of mRNA destabilization mechanisms, and the analysis of multiple mRNA localization datasets. We show that depletion of mRNA destabilization elements, such as m6A, AU-rich elements, and suboptimal codons, functions as a mechanism that mediates the localization of mRNAs associated with housekeeping functions to neurites in several types of neurons.
Link: Read Paper
The image analysis GitHub repo: GitHub link
Massively parallel identification of mRNA localization elements in primary cortical neurons
Authors: Samantha Mendonsa, Nicolai von Kügelgen, Sayaka Dantsuji, Maya Ron, Laura Breimann, Artem Baranovskii, Inga Lödige, Marieluise Kirchner, Meret Fischer, Nadja Zerna, Lucija Bujanic, Philipp Mertins, Igor Ulitsky & Marina Chekulaeva
Date: January 16, 2023
Journal: Nature Neuroscience
Abstract: Cells adopt highly polarized shapes and form distinct subcellular compartments in many cases due to the localization of many mRNAs to specific areas, where they are translated into proteins with local functions. This mRNA localization is mediated by specific cis-regulatory elements in mRNAs, commonly called ‘zipcodes’. Although there are hundreds of localized mRNAs, only a few zipcodes have been characterized. Here we describe a novel neuronal zipcode identification protocol (N-zip) that can identify zipcodes across hundreds of 3′ untranslated regions. This approach combines a method of separating the principal subcellular compartments of neurons—cell bodies and neurites—with a massively parallel reporter assay. N-zip identifies the let-7 binding site and (AU)n motif as de novo zipcodes in mouse primary cortical neurons. Our analysis also provides, to our knowledge, the first demonstration of an miRNA affecting mRNA localization and suggests a strategy for detecting many more zipcodes.
Link: Read Paper
The image analysis GitHub repo: GitHub link
RS-FISH: precise, interactive, fast, and scalable FISH spot detection
Authors: Ella Bahry *, Laura Breimann *, Marwan Zouinkhi *, Leo Epstein, Klim Kolyvanov, Nicholas Mamrak, Benjamin King, Xi Long, Kyle I S Harrington, Timothée Lionnet, Stephan Preibisch
Date: December 19, 2022
Journal: Nature Methods
Abstract: Fluorescent in-situ hybridization (FISH)-based methods extract spatially resolved genetic and epigenetic information from biological samples by detecting fluorescent spots in microscopy images, an often challenging task. We present Radial Symmetry-FISH (RS-FISH), an accurate, fast, and user-friendly software for spot detection in two- and three-dimensional images. RS-FISH offers interactive parameter tuning and readily scales to large datasets and image volumes of cleared or expanded samples using distributed processing on workstations, clusters, or the cloud. RS-FISH maintains high detection accuracy and low localization error across a wide range of signal-to-noise ratios, a key feature for single-molecule FISH, spatial transcriptomics, or spatial genomics applications.
Link: Read Paper
The software GitHub repo: GitHub link
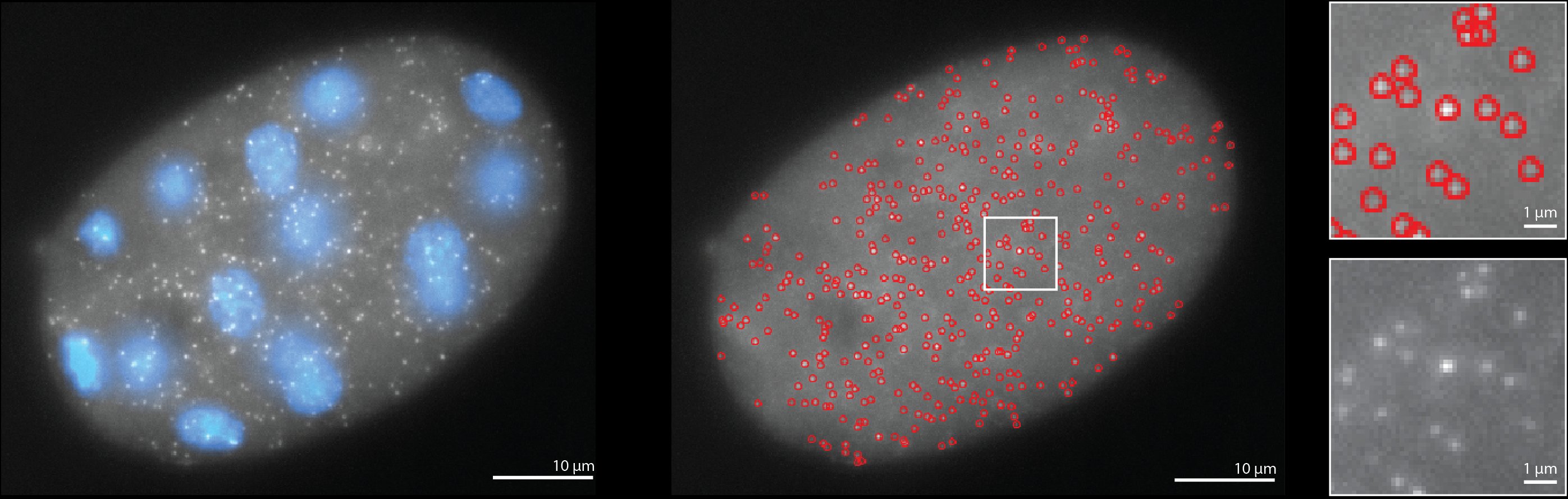
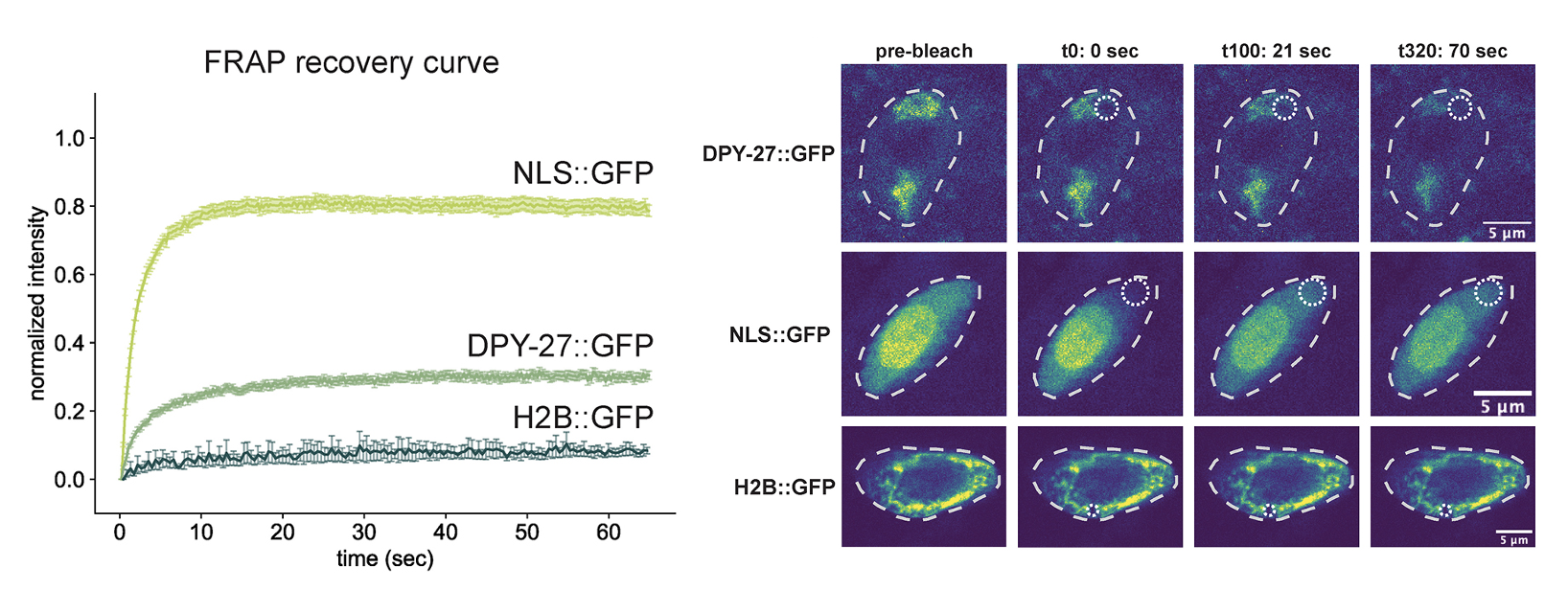
The histone H4 lysine 20 demethylase DPY-21 regulates the dynamics of condensin DC binding
Authors: Laura Breimann*#, Ana Karina Morao*, Jun Kim, David Sebastian Jimenez, Nina Maryn, Krishna Bikkasani, Michael J. Carrozza, Sarah E. Albritton, Maxwell Kramer, Lena Annika Street, Kustrim Cerimi, Vic-Fabienne Schumann, Ella Bahry, Stephan Preibisch, Andrew Woehler, and Sevinç Ercan#
Date: January 15, 2022
Journal: Journal of Cell Science
Abstract: Condensin is a multi-subunit structural maintenance of chromosomes (SMC) complex that binds to and compacts chromosomes. Here, we addressed the regulation of condensin binding dynamics using Caenorhabditis elegans condensin DC, which represses X chromosomes in hermaphrodites for dosage compensation. We established fluorescence recovery after photobleaching (FRAP) using the SMC4 homolog DPY-27 and showed that a well-characterized ATPase mutation abolishes DPY-27 binding to X chromosomes. Next, we performed FRAP in the background of several chromatin modifier mutants that cause varying degrees of X chromosome derepression. The greatest effect was in a null mutant of the H4K20me2 demethylase DPY-21, where the mobile fraction of condensin DC reduced from ∼30% to 10%. In contrast, a catalytic mutant of dpy-21 did not regulate condensin DC mobility. Hi-C sequencing data from the dpy-21 null mutant showed little change compared to wild-type data, uncoupling Hi-C-measured long-range DNA contacts from transcriptional repression of the X chromosomes. Taken together, our results indicate that DPY-21 has a non-catalytic role in regulating the dynamics of condensin DC binding, which is important for transcription repression.
Link: Read Paper
The analysis GitHub repo: GitHub link
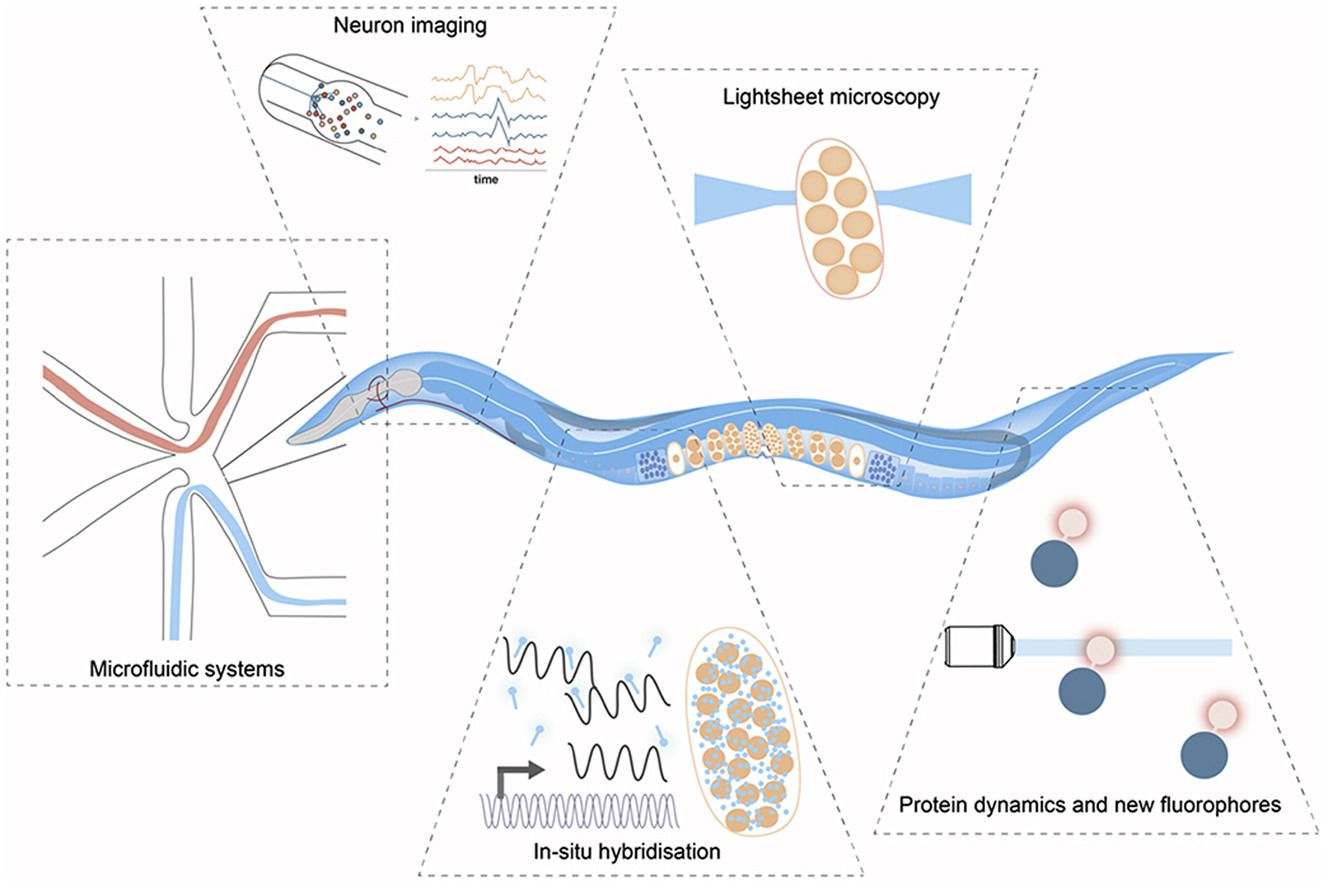
Light-microscopy methods in C. elegans research
Authors: Laura Breimann*, Friedrich Preusser*, Stephan Preibisch
Date: Feburary 1, 2019
Journal: Current Opinion in Systems Biology
Abstract: Ever since Caenorhabditis elegans was introduced as a model system it has been tightly linked to microscopy, which has led to significant advances in understanding biology over the last decades. Developing new technologies therefore is an essential part in the endeavor to gain further mechanistic insights into developmental biology. This review will discuss state-of-the-art developments in quantitative light microscopy in the context of C. elegans research as well as the impact these technologies have on the field. We will highlight future developments that currently promise to revolutionize biological research by combining sequencing-based single-cell technologies with high-resolution quantitative imaging.
Link: Read Paper
DamID profiling of dynamic Polycomb-binding sites in Drosophila imaginal disc development and tumorigenesis
Authors: Marco La Fortezza, Giovanna Grigolon, Andrea Cosolo, Alexey Pindyurin, Laura Breimann, Helmut Blum, Bas van Steensel & Anne-Kathrin Classen
Date: June 5, 2018
Journal: Epigenetics & Chromatin
Abstract: Background: Tracking dynamic protein–chromatin interactions in vivo is key to unravel transcriptional and epige‑netic transitions in development and disease. However, limited availability and heterogeneous tissue composition ofin vivo source material impose challenges on many experimental approaches.Results: Here we adapt cell‑type‑specific DamID‑seq profiling for use in Drosophila imaginal discs and make FLP/FRT‑based induction accessible to GAL driver‑mediated targeting of specific cell lineages. In a proof‑of‑principleapproach, we utilize ubiquitous DamID expression to describe dynamic transitions of Polycomb‑binding sites duringwing imaginal disc development and in a scrib tumorigenesis model. We identify Atf3 and Ets21C as novel Polycombtarget genes involved in scrib tumorigenesis and suggest that target gene regulation by Atf3 and AP‑1 transcriptionfactors, as well as modulation of insulator function, plays crucial roles in dynamic Polycomb‑binding at target sites. Weestablish these findings by DamID‑seq analysis of wing imaginal disc samples derived from 10 larvae.Conclusions: Our study opens avenues for robust profiling of small cell population in imaginal discs in vivo and pro‑vides insights into epigenetic changes underlying transcriptional responses to tumorigenic transformation.
Link: Read Paper